Therapeutic Peptides: Modulation of Protein-Protein Interactions
Contact:
pierre.tuffery _at_ u-paris.fr
The team’s main objective is the development of therapeutic peptides targeting protein-protein interactions. To this end, it brings together multidisciplinary skills combining computational modeling, chemistry, in vitro approaches, in order to (i) develop methodologies for the identification and optimization of candidate peptides, allowing their development from design to validation. proof of concept, and (ii) apply these developments in targeted pathological applications.
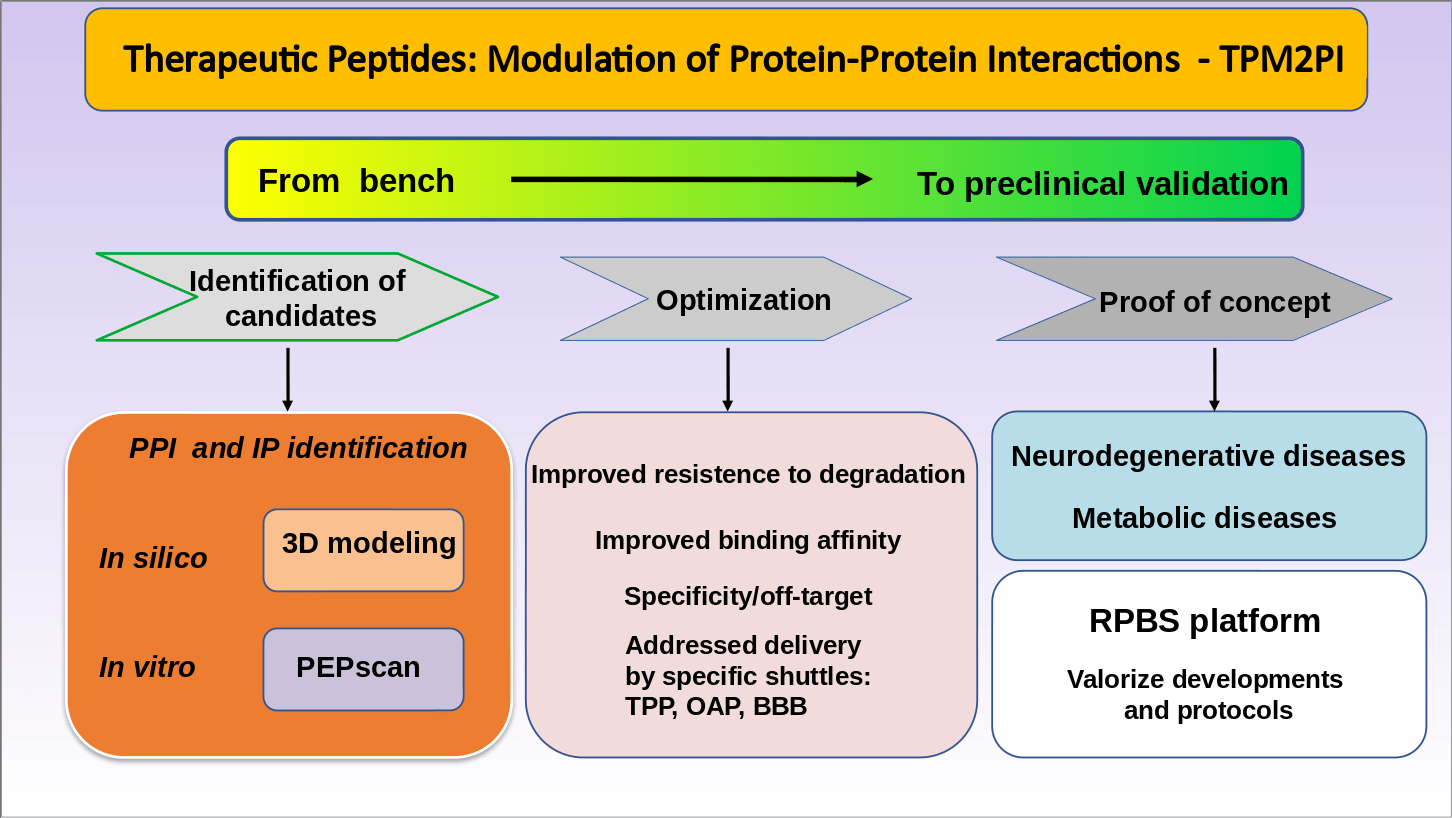
Research themes of the “TPM2PI” team
The identification of peptides by in silico and/or in vitro approaches is followed by an in silico and/or experimental optimization phase until the validation of the proof of concept in a pathological context.
© Copyright of the BFA Unit
Research themes
Prediction of the structure of peptides and their interactions with proteins
The team is interested in the development of in silico protocols for the identification of interfering peptides, in particular to target protein-protein interactions. To this end, she is interested in predicting the structure of peptides (PEP-FOLD – Rey et al., Nucleic Acids Res. 2023 doi: 10.1093/nar/gkad376.), and their interactions with protein targets, in terms of identification of interaction modes (peptide-protein complexes – de Vries et al., Nucleic Acids Res. 2017, doi: 10.1093/nar/gkx335.), and their stability (molecular dynamics simulation).
Peptide optimization
For the optimization of candidate peptides, we are interested in their optimization by cyclization (stabilité conformationnelle – et al., J Chem Inf Model. 2023, doi: 10.1021/acs.jcim.3c00865. ; et al., J. Chem. Inf. Model. 2018, doi: 10.1021/acs.jcim.8b00375 ) or in terms of specificity of interactions (off-target) (Rey et al., Nucleic Acids Res. 2019, doi: 10.1093/nar/gkz478.; , J Chem Inf Model. 2017, doi: 10.1021/acs.jcim.6b00529.).
Development of Interfering Peptides
Plusieurs projets de développement de peptides sont en cours, parmi lesquels:
- Maladie de Parkinson: identification d’un peptide ciblant l’interaction LRRK2/PP1 (Dong et al., PlosONE, 2020, doi: 10.1371/journal.pone.0237110. eCollection 2020.)
- Hépatocarcinome: peptides ciblant les cellules tumorales (Marin et al., Pharmaceutics, 2023, doi: 10.3390/pharmaceutics15041180. ; et al., Pharmaceutics, 2022, doi: 10.3390/pharmaceutics14102055.; Savier et al., Methods Mol Biol. 2022, doi: 10.1007/978-1-0716-1752-6_26.).
Functional characterization of peptides
- Syndrome de Down.
- Contrôle du métabolisme énergétique: recherche de modulateurs de récepteurs du système nerveux central pour l’exploration fine de la régulation du métabolisme.
RPBS platform (Ressource Parisienne en Bioinformatique Structurale)
The team hosts the RPBS structural bioinformatics platform (bioserv.rpbs.univ-paris-diderot.fr), a member of the French Institute of Bioinformatics (IFB). The platform provides a computing infrastructure dedicated to bioinformatics and online services (mobyle2.rpbs.univ-paris-diderot.fr). She is also available for advice, training in structural bioinformatics (analysis and modeling of the structure and function of proteins), collaborative or delivery projects.


Recent publications (selection)
- A Machine Learning Approach to Identify Key Residues Involved in Protein-Protein Interactions Exemplified with SARS-CoV-2 Variants.
Quitté L, Leclercq M, Prunier J, Scott-Boyer MP, Moroy G, Droit A. Int J Mol Sci. 2024 Jun 13;25(12):6535. doi: 10.3390/ijms25126535. PMID: 38928241 - Exploring a Structural Data Mining Approach to Design Linkers for Head-to-Tail Peptide Cyclization.
Karami Y, Murail S, Giribaldi J, Lefranc B, Defontaine F, Lesouhaitier O, Leprince J, de Vries S, Tufféry P. J Chem Inf Model. 2023 Oct 23;63(20):6436-6450. doi: 10.1021/acs.jcim.3c00865. Epub 2023 Oct 12. PMID: 37827517 - PEP-FOLD4: a pH-dependent force field for peptide structure prediction in aqueous solution.
Rey J, Murail S, de Vries S, Derreumaux P, Tuffery P. Nucleic Acids Res. 2023 Jul 5;51(W1):W432-W437. doi: 10.1093/nar/gkad376. PMID: 37166962 - A refined pH-dependent coarse-grained model for peptide structure prediction in aqueous solution.
Tufféry P, Derreumaux P. Front Bioinform. 2023 Jan 16;3:1113928. doi: 10.3389/fbinf.2023.1113928. eCollection 2023. PMID: 36727106
Team News
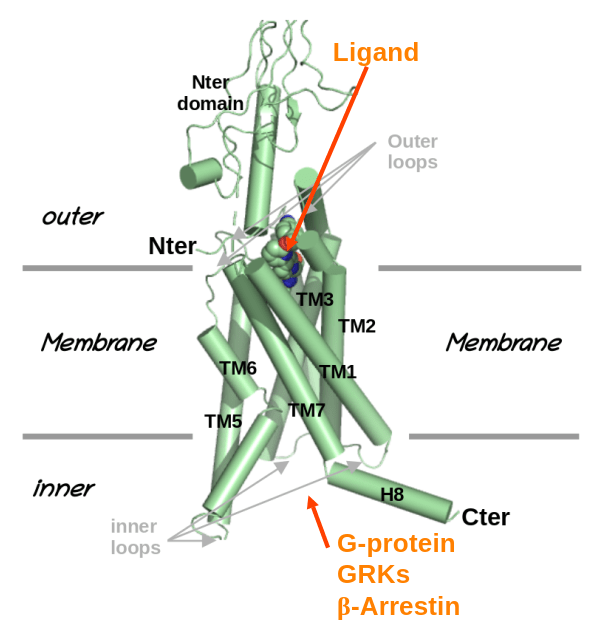
Welcome to Alaa Reguei, new PhD student at TPM2PI.
Alaa Reguei will join the TPM2PI team by october 2025, thank to a grant of thee graduate school "Drug Development". His work will focus on "Conception/optimisation of GPCRs biased modulators, combining AI and molecular simulation".
InIdex MetaboBrain
Acceptation du projet InIdex MetaboBrain, auquel l'équipe contribuera.
last update: 2025/02/11
